陈柱成实验室
Chen Zhucheng Laboratory
We use advanced tools in structural biology, such as crystallography and cryo-electron microscopy, in combination with biochemistry, genetics and other methods, to analyze chromatin-related molecular machines.
Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis
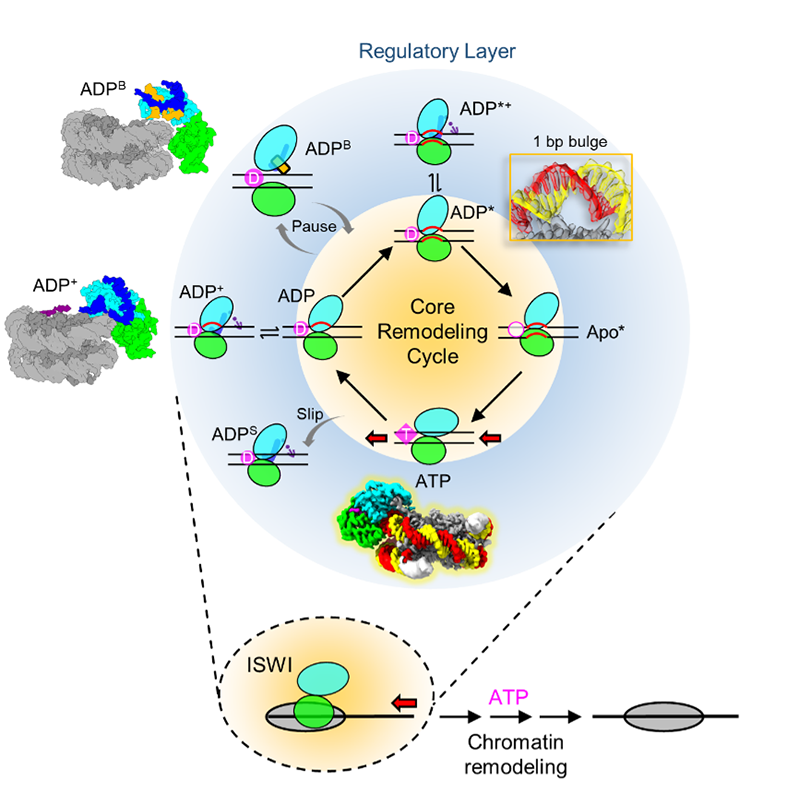
Chromatin remodelers use energy from ATP hydrolysis to reposite nucleosomes around genomic DNA sites and remodel chromatin. They play important roles in shaping chromatin structures and controlling gene expression, cell fate determination, and human diseases. How chromatin remodelers use the energy of ATP hydrolysis to promote nucleosome sliding is a puzzling question.
Chromatin remodeling is a multi-step dynamic process coupled with ATP hydrolysis. The main challenge to the mechanisms of chromatin remodeling concerns how the remodeler enzymes overcome the extensive histone–DNA interactions to slide the nucleosome. To tease these mechanisms apart, it was necessary to establish how chromatin remodelers interact with the nucleosome. In 2016, we determined the crystal structures of Snf2 and ISWI motor domains. In 2017, we determined the cryoEM structure of the motor domains of Snf2 bound to the nucleosome in the nucleotide-free, apo state, which provided the first glimpse into the motor-nucleosome interaction. In 2019, we then determined the structures of Snf2 and ISWI bound to the nucleosome in the presence of ADP and ADP-BeFx. Together, our findings suggest a fundamental mechanism for the DNA translocation that underlies chromatin remodelling.
In 2025, we report structures of imitation switch (ISWI) bound to the nucleosome during active ATP hydrolysis and remodeling, revealing conformational transitions of the remodeling motor across the adenosine triphosphatase (ATPase) cycle. The DNA strands are distorted accordingly, showing one full base-pair bulge and a loss of histone contact at the site of motor binding in the adenosine diphosphate* b and Apo* states.

扫一扫关注微信